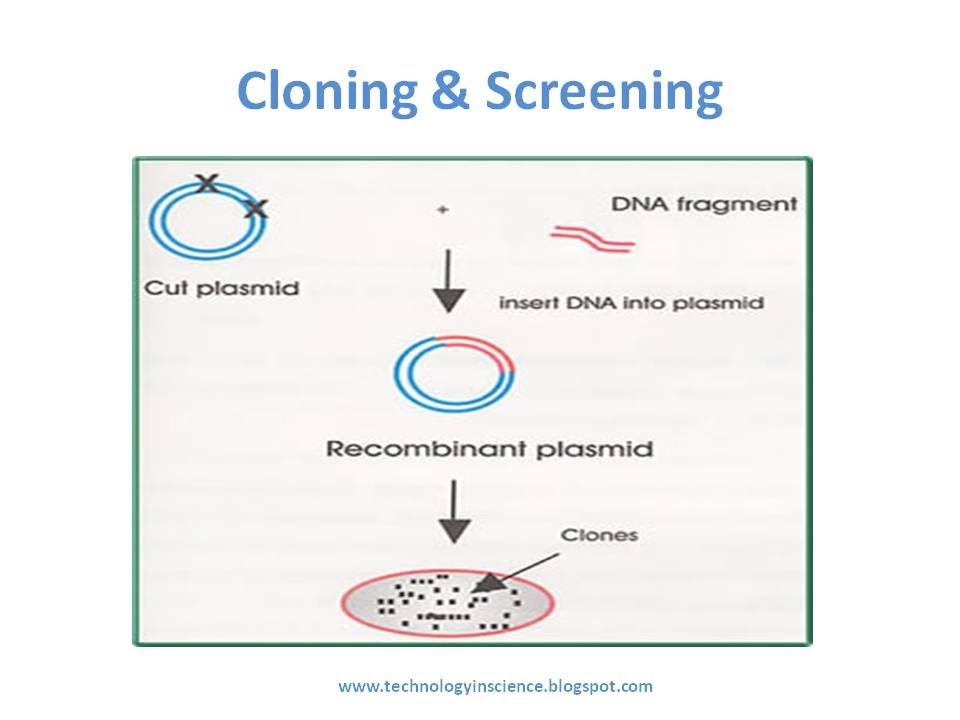
Introduction
Escherichia coli Recombinants is one of the organisms of choice for the production of recombinant proteins. Its use as a cell factory is well established and has become the most popular expression platform. For this reason, there are many molecular tools and protocols available for the high-level production of heterologous proteins, such as a wide catalogue of expression plasmids, a large number of engineered strains, and many breeding strategies. We review the different approaches to recombinant protein synthesis in E. coli and discuss recent progress in this ever-growing field.
Keywords: expression vectors, secretion, molecular chaperones.
DNA sequences involved in transcription.
Three different DNA sequences and one multi-component protein are involved in gene transcription: (1) promoter, (2) transcriptional terminator, (3) regulatory sequence, and (4) RNA polymerase. RNA polymerase consists of five different components called a, b, B, w and s. While a2bbw constitutes the core enzyme, the addition of s which confers promoter specificity constitutes the holoenzyme. The N-terminal part of a is involved in the formation of the dimer and the union ab and b’, and its C-terminus, linked by a flexible linker to its N-terminus, is responsible for the interaction with the UP element present upstream of some promoters (see below) or with some transcriptional activators.
The b subunit binds to dNTPs, contains the catalytic domain, and is the target of the antibiotic rifampin, while b’ allows nonspecific DNA binding. The role of w is largely unknown, but it is hypothesized that it plays a role in RNA polymerase assembly. Although all bacterial species analyzed so far contain only one gene, each of which encodes the core enzyme components, most species possess genes encoding multiple s-factors. One of these factors functions as a primary or housekeeping factor and is involved in the transcription of all genes necessary for growth during the vegetative phase.
Additional s-factors are called secondary or alternative s-factors and are only needed under specific growth conditions (Gruber and Gross, 2003). E. coli encodes six alternative factors in which s32 is needed after a temperature surge and sS replaces the housekeeping factor s70 during the stationary phase. Until now, only s70 is used in the production of recombinant proteins.
As mentioned above, the s-factor is responsible for promoter recognition, and it follows that each s-factor recognizes a different promoter. Promoters normally consist of three regions called box -35 and -10 and the spacer region that separates both boxes. The alignment of many promoters allows the deduction of a so-called consensus sequence, and the consensus sequence for s70 is TTGACA-N17-TATA. This sequence represents the optimal promoter sequence with a 17 nucleotide spacer region.
It should be noted that there is not a single promoter present on the E. coli chromosome identical to the consensus sequence. In most cases, there are one or two deviations in both the -35 and -10 boxes. Additionally, some promoters contain a fourth region, the UP element located upstream of the -35 box. The UP element consists of an AT-rich sequence that allows interaction with the C-terminal domain of the subunit, increasing the strength of the promoter. None of the promoters that drive the production of recombinant proteins uses the UP element.
DNA sequences involved in translation.
Due to the complexity of the process, the determinants of the initiation of protein synthesis have been difficult to decipher. It became clear that the wide range of translational efficiencies of different mRNAs is predominantly due to the structure at the 5′ end of each mRNA species. Therefore, no universal sequence for the efficient initiation of translation has been devised. The translation start region comprises four different sequences: (1) the Shine-Dalgarno sequence, (2) the start codon, (3) the spacer region between the Shine-Dalgarno sequence and the start codon, and (4) sometimes translation enhancers.
Furthermore, the secondary structure in the translation initiation region of the mRNA plays an important role in the efficiency of gene expression. Occlusion of the Shine-Dalgarno sequence and/or the start codon by a stem-loop structure has been shown to prevent access to the 30S ribosomal subunit and inhibit translation (Ramesh et al., 1994).
There are two reported cases in which this principle is used to significantly reduce downstream translation, namely upon mRNA encoding the s32 heat shock sigma factor in E. coli and mRNAs encoding small proteins of heat shock in rhizobia (Morita et al., 1999; Nocker et al., 2001). In both cases, translation of these mRNAs is achieved under heat shock conditions leading to secondary structure melting. There are possibilities to minimize the secondary structure of the mRNA in the translation initiation region.
Whereas enrichment of the RBS with adenine and thymine residues enhanced the expression of certain genes (Chen et al., 1994), mutation of specific nucleotides upstream or downstream of the Shine-Dalgarno sequence suppressed the formation of secondary structures of RBS. mRNA and improved translation efficiency (Coleman et al., 1985; Gross et al., 1990). Sequences have been identified that markedly enhance the expression of recombinant genes, and these modules have been termed translational enhancers. An example is a U-rich region immediately upstream of the Shine-Dalgarno sequence in the E. coli at gene (McCarthy et al., 1985). This 30 base sequence has been used successfully to overexpress the human interleukin-2 and interferon-beta genes (McCarthy et al., 1986).