Description
To comprehensively analyze bacterial gene function, it is important to simultaneously generate multiple genetic modifications within the target gene. However, current genetic engineering approaches, primarily using suicide vectors or λ-red homologous recombination-based systems, are tedious and technically difficult to perform. Here, we develop a flexible and easy method to simultaneously construct multiple modifications at the same locus on the chromosome of Salmonella Typhimurium Recombinant.
The method combines an efficient in vitro assembly system, in vivo red homologous recombination, and a sacB counterselection marker. To test this approach, several modification fragments for the cpxR, CPA and acrB target genes were quickly and efficiently constructed in vitro with the continuous assembly system. SacBKan cassettes generated via polymerase chain reaction were inserted into target loci in the genome of Salmonella Typhimurium strain CVCC541.
The resulting kanamycin-resistant recombinants containing pKD46 were selected and used as intermediate strains. Multiple target gene modifications were then carried out simultaneously through allelic swapping using various homologous recombinogenic DNA fragments to replace the sack and cassettes on the chromosomes of the intermediate strains. With this method, we successfully carried out site-directed mutagenesis, perfect deletion, and 3 × FLAG tagging of target genes. This method can be used in any bacterial species that supports sacB gene activity and λ-red mediated recombination, allowing in-depth functional analysis of bacterial genes.
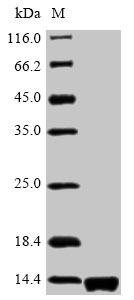
DNA manipulation
PrimeStar Max DNA polymerase (Takara Bio), with a mismatch ratio of 12 bases per 542,580 total bases, was selected to minimize errors in the PCR products. Genomic DNA fragments were purified with a MiniBEST bacterial genomic DNA extraction kit (Takara Bio) and MiniBEST agarose gel DNA extraction kit (Takara Bio), while plasmid DNA samples were purified with a MiniBEST plasmid purification kit (Takara Bio).
PCR assays were performed using PrimeStar Max DNA Polymerase (Takara Bio). Restriction and T4 DNA ligase enzymes were purchased from TransGen Biotech. In vitro, seamless assembly of different DNA fragments was performed using a pEASY-Uni seamless assembly and cloning kit (TransGen Biotech) according to the manufacturer’s instructions.
Construction of the psacBKan plasmid
The Kan resistance cassette was amplified from pET-30a using primers Kan-BamHI-F and Kan-EcoRI-R (containing the 20 bp P2 sequence at the 5′ end), and the gel-purified product was digested with EcoRI and BamHI. The sacB gene cassette (the negative selection marker) was then amplified from plasmid pUC57-sacB using the primers such-F-SphI (containing the 20 bp P1 sequence at the 5′ end) and sacB-R- BamHI, and the gel-purified product was digested with SphI and BamHI.
The two cassettes were then ligated into pSTV28 double digested with EcoRI and SphI using T4 DNA ligase, and the resulting recombinant plasmid, designated psacBKan, was transformed into chemically competent phage-resistant E. coli Trans1-T1 (TransGen Biotech).
To confirm that the sack and cassette could be used for selection, psacBKan was electroporated into the wild-type strain JS. After overnight growth at 37°C, wild-type and psacBKan-carrying strains were diluted 1:100 in LB medium and grown at 37°C to log phase. Aliquots (100 µl) of each culture were then spread onto LB agar plates containing 8% sucrose or kanamycin (50 µg/ml) and incubated at 37°C for 24 h.
Simultaneous generation of multiple genetic mutations
Competent cells prepared for the intermediate sacBKan-containing strains harbouring pKD46 were induced with L-arabinose. Aliquots (200 ng) of the different recombinogenic DNA fragments were then electroporated into the corresponding intermediate strains. After electroporation, cells were recovered in 1 ml of SOC medium at 37 °C, 200 rpm, for 2 h. Serial dilutions of the bacterial cultures were then spread on NaCl minus LB agar plates supplemented with 8% (w/v) sucrose and incubated at 37°C for 24 h to select a sucrose-resistant strain.
Correct mutations were confirmed by PCR using locus-specific primers. The recombination efficiencies of the different recombinogenic DNA fragments were evaluated based on the proportion of colonies with correct PCR identification. Finally, the accuracy rate of the genetic modifications was further verified by Sanger sequencing.

Immunodetection analysis
Immunodetection assays were carried out as previously described (Uzzau et al., 2001). Briefly, strains carrying 3 × FLAG fusion-tagged genes were grown in LB medium to early stationary phase. After centrifugation (10,000 × g, 5 min), bacterial pellets from 1 ml culture were resuspended in 100 μl phosphate-buffered saline and mixed with protein loading buffer before boiling for 10 min and then place on ice.
The resulting lysates were resolved by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, blocked with Tris-buffered saline (pH 8.0) and 3% skim milk. (v/v), were tested with anti-FLAG M2. monoclonal antibodies (mAb) (1:1,000) (Sigma), and then incubated with a goat anti-rabbit IgG (whole molecule) horseradish peroxidase (HRP) conjugate (1:4,000). FLAG-tagged proteins were visualized using enhanced chemiluminescence.
Drug Susceptibility Assay
Minimum inhibitory concentrations (MICs) of selected antibiotics against all strains were determined using the two-broth microdilution method according to the Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute, 2008, 2012 ). The MICs of amikacin (AMK) and cefuroxime (CMX) were determined independently at least three times.